Dump and reload example¶
This notebook illustrates how to dump a cancer simulation to disk, reload it, and continue with changed parameters. This feature allows to model different stages of tumour growth characterized by varying growth parameters (e.g. death_probability and adv_mutant_division_probability).
Import modules¶
[1]:
# The cancer simulation module.
from casim import casim
[2]:
# 3rd party modules.
import os, shutil
from wand.image import Image as WImage
from glob import glob
[3]:
import pickle
from matplotlib import pyplot
Define some utility functions¶
[4]:
def unpickle(sim, pkl):
with open(os.path.join(sim._CancerSimulator__simdir, pkl),'rb') as fp:
return pickle.load(fp)
[5]:
def clear_outdir(dir):
if os.path.isdir(dir):
shutil.rmtree(dir)
Setup parameters¶
Initially, cells with advantageous mutations divide only seldomly, the tumour is dormant.
[22]:
parameters=casim.CancerSimulatorParameters(
matrix_size=200,
number_of_generations=50,
division_probability=0.6,
adv_mutant_division_probability=0.5,
death_probability=0.3,
adv_mutant_death_probability=0.05,
mutation_probability=0.5,
adv_mutant_mutation_probability=1.0,
number_of_mutations_per_division=10,
adv_mutation_wait_time=40,
number_of_initial_mutations=2,
sampling_fraction=0.9,
plot_tumour_growth=True,
export_tumour=True
)
Setup the simulation engine.¶
[23]:
outdir = 'out'
clear_outdir(outdir)
[24]:
cancer_sim = casim.CancerSimulator(parameters, seed=1, outdir='out/')
2020-08-19 12:29:15,983 INFO: Running in single tumour mode.
2020-08-19 12:29:15,985 INFO: First cell at (100, 100).
Run the simulation¶
[25]:
cancer_sim.run()
2020-08-19 12:29:16,398 INFO: Ready to start CancerSim run with these parameters:
2020-08-19 12:29:16,400 INFO: matrix_size = 200
2020-08-19 12:29:16,401 INFO: number_of_generations = 50
2020-08-19 12:29:16,403 INFO: division_probability = 0.6
2020-08-19 12:29:16,404 INFO: adv_mutant_division_probability = 0.5
2020-08-19 12:29:16,405 INFO: death_probability = 0.3
2020-08-19 12:29:16,406 INFO: adv_mutant_death_probability = 0.05
2020-08-19 12:29:16,407 INFO: mutation_probability = 0.5
2020-08-19 12:29:16,409 INFO: adv_mutant_mutation_probability = 1.0
2020-08-19 12:29:16,410 INFO: number_of_mutations_per_division = 10
2020-08-19 12:29:16,411 INFO: adv_mutation_wait_time = 40
2020-08-19 12:29:16,412 INFO: number_of_initial_mutations = 2
2020-08-19 12:29:16,413 INFO: tumour_multiplicity = single
2020-08-19 12:29:16,414 INFO: read_depth = 100
2020-08-19 12:29:16,415 INFO: sampling_fraction = 0.9
2020-08-19 12:29:16,416 INFO: plot_tumour_growth = True
2020-08-19 12:29:16,418 INFO: export_tumour = True
2020-08-19 12:29:16,419 INFO: Tumour growth in progress.
2020-08-19 12:29:16,621 INFO: New beneficial mutation: 2076
2020-08-19 12:29:16,653 INFO: Division of beneficial mutation carrier. Cell index = (108, 110), mutation index = 2076, place_to_divide=(109, 111)
2020-08-19 12:29:16,675 INFO: Division of beneficial mutation carrier. Cell index = (108, 110), mutation index = 2076, place_to_divide=(108, 111)
2020-08-19 12:29:16,688 INFO: Division of beneficial mutation carrier. Cell index = (108, 111), mutation index = 2076, place_to_divide=(109, 110)
2020-08-19 12:29:16,738 INFO: Division of beneficial mutation carrier. Cell index = (109, 111), mutation index = 2076, place_to_divide=(110, 111)
2020-08-19 12:29:16,741 INFO: Division of beneficial mutation carrier. Cell index = (108, 111), mutation index = 2076, place_to_divide=(108, 112)
2020-08-19 12:29:16,757 INFO: Division of beneficial mutation carrier. Cell index = (110, 111), mutation index = 2076, place_to_divide=(110, 112)
2020-08-19 12:29:16,771 INFO: Division of beneficial mutation carrier. Cell index = (108, 111), mutation index = 2076, place_to_divide=(108, 112)
2020-08-19 12:29:16,772 INFO: Mutation of beneficial mutation carrier. Cell index = (108, 111), mutation index = 2076, place to divide = (108, 112)
2020-08-19 12:29:16,789 INFO: Division of beneficial mutation carrier. Cell index = (110, 111), mutation index = 2076, place_to_divide=(111, 112)
2020-08-19 12:29:16,790 INFO: Mutation of beneficial mutation carrier. Cell index = (110, 111), mutation index = 2076, place to divide = (111, 112)
2020-08-19 12:29:16,792 INFO: Division of beneficial mutation carrier. Cell index = (110, 112), mutation index = 2076, place_to_divide=(111, 113)
2020-08-19 12:29:16,793 INFO: Mutation of beneficial mutation carrier. Cell index = (110, 112), mutation index = 2076, place to divide = (111, 113)
2020-08-19 12:29:16,878 INFO: All generations finished. Starting tumour reconstruction.
2020-08-19 12:29:16,884 INFO: Reconstruction done, get statistics.
2020-08-19 12:29:17,206 INFO: Exporting simulation data
2020-08-19 12:29:17,443 INFO: Growth curve graph written to out/cancer_1/simOutput/growthCurve.pdf.
2020-08-19 12:29:18,021 INFO: CancerSim run has finished.
2020-08-19 12:29:18,023 INFO: Simulation output written to: out/cancer_1/simOutput.
2020-08-19 12:29:18,023 INFO: Log files written to: out/cancer_1/log.
2020-08-19 12:29:18,024 INFO: Consumed Wall time of this run: 1.601802 s.
[25]:
0
Visualize the first stage of the run¶
[26]:
pdfs = glob(os.path.join(cancer_sim.outdir,'cancer_1', 'simOutput',"*.pdf"))
[27]:
for pdf in pdfs:
print(pdf)
display(WImage(filename=pdf))
out/cancer_1/simOutput/sampleHistogram_97_107.pdf
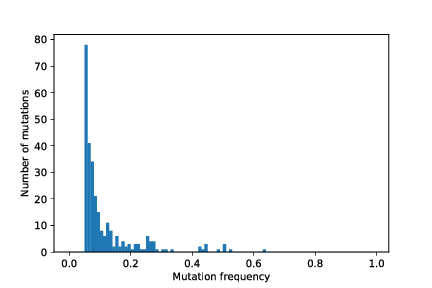
out/cancer_1/simOutput/growthCurve.pdf
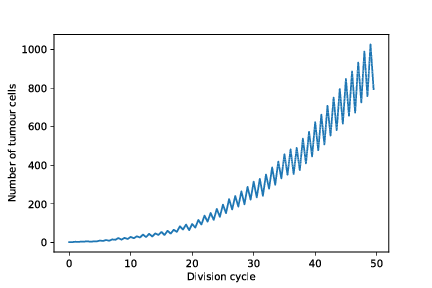
out/cancer_1/simOutput/wholeTumourVAFHistogram.pdf
Load the mutation matrix¶
[28]:
mtx=unpickle(cancer_sim,'mtx.p').toarray()
Plot the tumour as a 2D map color coding the mutation ID¶
[29]:
cmap = pyplot.cm.get_cmap('PRGn', mtx.max()+1)
pyplot.matshow(mtx, cmap=cmap)
pyplot.colorbar()
[29]:
<matplotlib.colorbar.Colorbar at 0x7f3798e37390>

Dump the simulation¶
[30]:
cancer_sim.dump()
Reload the simulation¶
[31]:
restart_with_mods = casim.load_cancer_simulation(cancer_sim.dumpfile)
Change adv. mutant properties and run for 20 more generations.¶
[32]:
restart_with_mods.parameters.adv_mutant_division_probability = 1.0
restart_with_mods.parameters.death_probability = 0.5
restart_with_mods.parameters.number_of_generations = 20
[33]:
outdir = 'restart_with_mods'
clear_outdir(outdir)
restart_with_mods.outdir = outdir
[34]:
restart_with_mods.run()
2020-08-19 12:29:32,477 INFO: Ready to start CancerSim run with these parameters:
2020-08-19 12:29:32,479 INFO: matrix_size = 200
2020-08-19 12:29:32,481 INFO: number_of_generations = 20
2020-08-19 12:29:32,482 INFO: division_probability = 0.6
2020-08-19 12:29:32,483 INFO: adv_mutant_division_probability = 1.0
2020-08-19 12:29:32,485 INFO: death_probability = 0.5
2020-08-19 12:29:32,486 INFO: adv_mutant_death_probability = 0.05
2020-08-19 12:29:32,487 INFO: mutation_probability = 0.5
2020-08-19 12:29:32,489 INFO: adv_mutant_mutation_probability = 1.0
2020-08-19 12:29:32,490 INFO: number_of_mutations_per_division = 10
2020-08-19 12:29:32,491 INFO: adv_mutation_wait_time = 40
2020-08-19 12:29:32,492 INFO: number_of_initial_mutations = 2
2020-08-19 12:29:32,494 INFO: tumour_multiplicity = single
2020-08-19 12:29:32,495 INFO: read_depth = 100
2020-08-19 12:29:32,496 INFO: sampling_fraction = 0.9
2020-08-19 12:29:32,497 INFO: plot_tumour_growth = True
2020-08-19 12:29:32,498 INFO: export_tumour = True
2020-08-19 12:29:32,499 INFO: Tumour growth in progress.
2020-08-19 12:29:33,198 INFO: All generations finished. Starting tumour reconstruction.
2020-08-19 12:29:33,209 INFO: Reconstruction done, get statistics.
2020-08-19 12:29:34,007 INFO: Exporting simulation data
2020-08-19 12:29:34,184 INFO: Growth curve graph written to restart_with_mods/cancer_1/simOutput/growthCurve.pdf.
2020-08-19 12:29:36,200 INFO: CancerSim run has finished.
2020-08-19 12:29:36,202 INFO: Simulation output written to: restart_with_mods/cancer_1/simOutput.
2020-08-19 12:29:36,202 INFO: Log files written to: restart_with_mods/cancer_1/log.
2020-08-19 12:29:36,203 INFO: Consumed Wall time of this run: 3.699930 s.
[34]:
0
Output¶
[35]:
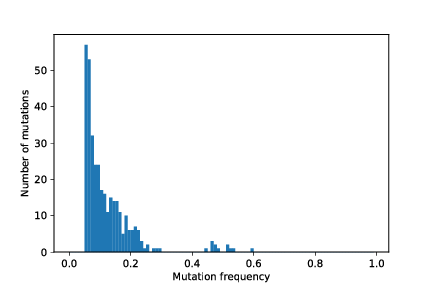
pdfs = glob(os.path.join(restart_with_mods.outdir,'cancer_1', 'simOutput',"*.pdf"))
for pdf in pdfs:
print(pdf)
display(WImage(filename=pdf))
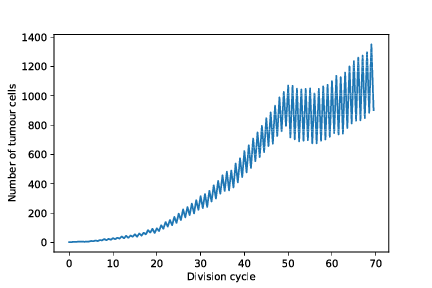
restart_with_mods/cancer_1/simOutput/growthCurve.pdf
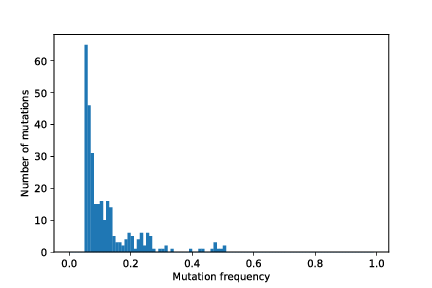
restart_with_mods/cancer_1/simOutput/wholeTumourVAFHistogram.pdf
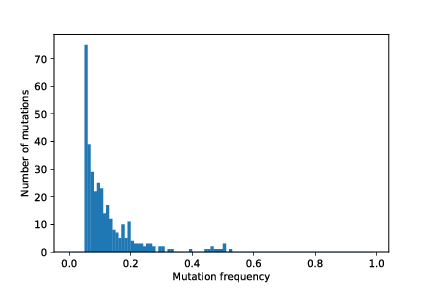
restart_with_mods/cancer_1/simOutput/sampleHistogram_86_78.pdf
[20]:
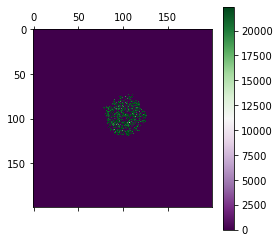
# Load the mutation matrix
mtx=unpickle(restart_with_mods, 'mtx.p').toarray()
[21]:
# Plot the tumour as a 2D map color coding the mutation ID
cmap = pyplot.cm.get_cmap('PRGn', mtx.max()+1)
pyplot.matshow(mtx, cmap=cmap)
pyplot.colorbar()
[21]:
<matplotlib.colorbar.Colorbar at 0x7f37961dc668>
[ ]: