Quickstart example for cancer simulation¶
This notebook illustrates how to run a 2D cancer simulation using CancerSim. It’s a rather contrieved example with no actual scientific background. It’s main purpose is to give a simple example to be used as a template for more complex simulation runs. It also demonstrates how to parametrize the simulation through the CancerSimulationParameters API instead of the params.py module.
The simulation treats a 2D tumour on a 20x20 grid for 10 generations. Healthy cells and cancer cells differ by their division probabilities (0.5 vs 0.8), death probabilities (0.1 vs. 0.4), and mutation probabilities (0.2 vs. 0.8). The first cancer cell carries 2 mutations, new mutations occur with a 20% probability and 10 fold abundance. The first advantageous mutation occurs in the 3rd generation. We sample 10% of the tumour at a read depth of 100.
After the run, we plot the tumour growth curve and the mutation frequency histograms for the whole tumour and the sampled tumour.
Import modules¶
[6]:
# The cancer simulation module.
from casim import casim
import logging
[2]:
# 3rd party modules.
import os, shutil
from wand.image import Image as WImage # To render pdf images in nb.
from glob import glob
Setup parameters¶
[3]:
parameters=casim.CancerSimulatorParameters(
matrix_size=20,
number_of_generations=10,
division_probability=0.5,
adv_mutant_division_probability=0.8,
death_probability=0.1,
adv_mutant_death_probability=0.4,
mutation_probability=0.2,
adv_mutant_mutation_probability=0.8,
number_of_mutations_per_division=10,
adv_mutation_wait_time=3,
number_of_initial_mutations=2,
sampling_fraction=0.1,
plot_tumour_growth=True,
export_tumour=True
)
Setup the simulation engine.¶
[4]:
if os.path.isdir('out'):
shutil.rmtree('out')
[7]:
cancer_sim = casim.CancerSimulator(parameters, seed=1, outdir='out/')
[11]:
# Get more verbose logging.
logger = logging.getLogger().setLevel("INFO")
Run the simulation¶
[10]:
cancer_sim.run()
2020-08-10 16:14:45,538 INFO: Running in single tumour mode.
2020-08-10 16:14:45,540 INFO: First cell at (10, 10).
2020-08-10 16:14:45,542 INFO: Ready to start CancerSim run with these parameters:
2020-08-10 16:14:45,543 INFO: matrix_size = 20
2020-08-10 16:14:45,544 INFO: number_of_generations = 10
2020-08-10 16:14:45,545 INFO: division_probability = 0.5
2020-08-10 16:14:45,546 INFO: adv_mutant_division_probability = 0.8
2020-08-10 16:14:45,547 INFO: death_probability = 0.1
2020-08-10 16:14:45,547 INFO: adv_mutant_death_probability = 0.4
2020-08-10 16:14:45,548 INFO: mutation_probability = 0.2
2020-08-10 16:14:45,549 INFO: adv_mutant_mutation_probability = 0.8
2020-08-10 16:14:45,550 INFO: number_of_mutations_per_division = 10
2020-08-10 16:14:45,551 INFO: adv_mutation_wait_time = 3
2020-08-10 16:14:45,551 INFO: number_of_initial_mutations = 2
2020-08-10 16:14:45,552 INFO: tumour_multiplicity = single
2020-08-10 16:14:45,553 INFO: read_depth = 100
2020-08-10 16:14:45,554 INFO: sampling_fraction = 0.1
2020-08-10 16:14:45,554 INFO: plot_tumour_growth = True
2020-08-10 16:14:45,555 INFO: export_tumour = True
2020-08-10 16:14:45,556 INFO: Tumour growth in progress.
2020-08-10 16:14:45,560 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,567 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,571 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,572 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,578 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,582 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,583 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,586 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,587 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,588 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,591 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,593 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,593 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,594 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,595 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,598 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,598 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,602 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,604 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,605 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,607 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,609 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,610 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,612 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,616 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,616 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,617 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,618 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,619 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,621 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,622 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,623 INFO: No new mutation in normal division, inheriting from parent
2020-08-10 16:14:45,624 INFO: All generations finished. Starting tumour reconstruction.
2020-08-10 16:14:45,625 INFO: Reconstruction done, get statistics.
2020-08-10 16:14:45,630 INFO: Exporting simulation data
2020-08-10 16:14:45,901 INFO: Growth curve graph written to out/cancer_1/simOutput/growthCurve.pdf.
2020-08-10 16:14:46,082 INFO: CancerSim run has finished.
2020-08-10 16:14:46,084 INFO: Simulation output written to: out/cancer_1/simOutput.
2020-08-10 16:14:46,084 INFO: Log files written to: out/cancer_1/log.
2020-08-10 16:14:46,085 INFO: Consumed Wall time of this run: 0.525742 s.
[10]:
0
<Figure size 432x288 with 0 Axes>
Output¶
After the run has finished, you should find the results in out/cancer_1/simOutput.
[12]:
!ls out/cancer_1/simOutput
death_list.p mtx_VAF.txt sample_out_12_11.txt
growthCurve.pdf mut_container.p wholeTumourVAFHistogram.pdf
mtx.p sampleHistogram_12_11.pdf
Files with the extension .p are binary files (python pickles) needed to restart a simulation. Let’s take a look at the .txt files. They contain the simulation output: mtx_VAF.txt is a datafile with three columns: mutation_id lists the index of each primary mutation, additional_mut_id indexes the subsequent mutations that occur in a cell of a given mutation_id; frequency is the frequency at which a given mutation occurs.
The file sample_out_502_488.txt lists all mutations of the artificial sample taken from the whole tumour. Columns are identical to mtx_VAF.txt.
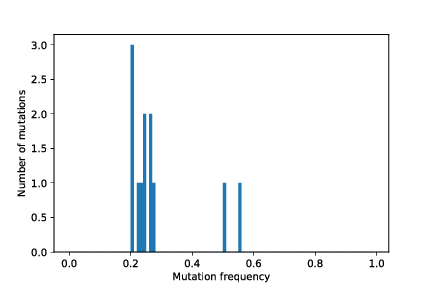
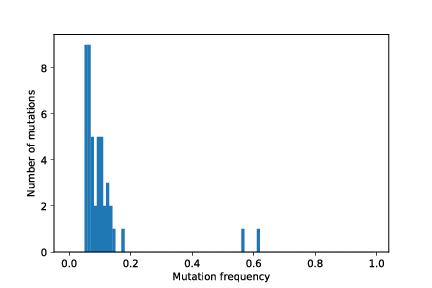
The two .pdf files are plots of the whole tumour histogram and the sampled tumour histogram, respectively. You should see figures similar to these:
Display results.¶
The mutation frequency histogram for the whole tumour and the for sampled part of the tumour, as well as the tumour growth curve (number of tumour cells vs. time) are stored as pdf images in the simulation output directory.
[13]:
image_path=os.path.join(cancer_sim.outdir,'cancer_1', 'simOutput')
[14]:
pdfs = glob(os.path.join(image_path,"*.pdf"))
[15]:
pdfs
[15]:
['out/cancer_1/simOutput/growthCurve.pdf',
'out/cancer_1/simOutput/sampleHistogram_12_11.pdf',
'out/cancer_1/simOutput/wholeTumourVAFHistogram.pdf']
[16]:
for pdf in pdfs:
print(pdf)
display(WImage(filename=pdf))
out/cancer_1/simOutput/growthCurve.pdf
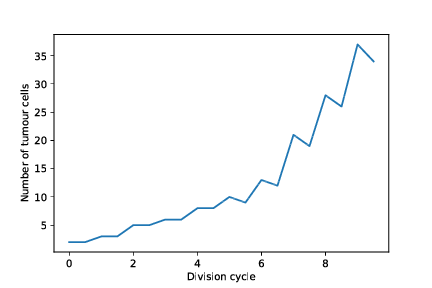
out/cancer_1/simOutput/sampleHistogram_12_11.pdf
out/cancer_1/simOutput/wholeTumourVAFHistogram.pdf
Load binary files (pickles)¶
The remaining output files are serialized versions (“pickles”) of the tumour geometry as a 2D matrix (mtx.p), the death list (death_list.p), and the mutation list (list of tuples listing the parent and the mutation ID of each tumour cell, mut_container.p).
To read the pickled data, we define a utility function
[17]:
import pickle
from matplotlib import pyplot
[18]:
def unpickle(file):
with open(os.path.join(cancer_sim._CancerSimulator__simdir, file),'rb') as fp:
return pickle.load(fp)
[19]:
# Load the mutation matrix
mtx=unpickle('mtx.p').toarray()
[20]:
# Plot the tumour as a 2D map color coding the mutation ID
cmap = pyplot.cm.get_cmap('PRGn', mtx.max()+1)
pyplot.matshow(mtx, cmap=cmap)
pyplot.colorbar()
[20]:
<matplotlib.colorbar.Colorbar at 0x7fc24be3b358>

[21]:
unpickle('mut_container.p')
[21]:
[(0, 0),
(0, 1),
(1, 2),
(1, 3),
(1, 4),
(1, 5),
(5, 6),
(5, 7),
(6, 8),
(6, 9),
(1, 10),
(1, 11),
(3, 12),
(3, 13),
(3, 14),
(3, 15),
(4, 16),
(4, 17),
(15, 18),
(15, 19),
(1, 20),
(1, 21)]